Tag: Isoform
-
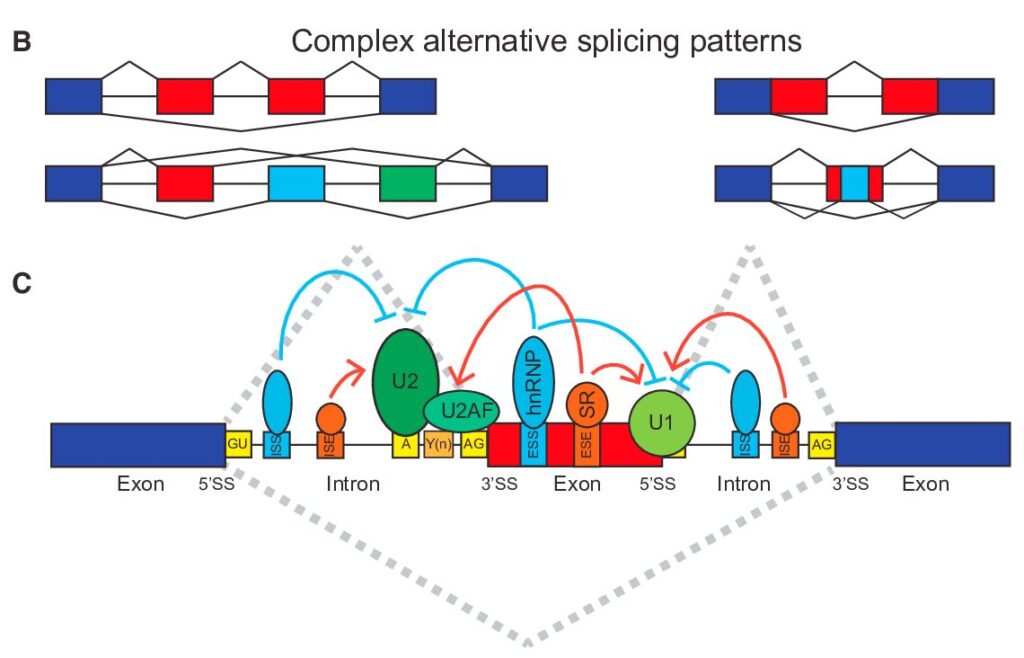
How to Analyze RNAseq Data for Absolute Beginners Part 11: Mastering Transcript-Level Alternative Splicing Analysis
From Gene-Level to Transcript-Level Analysis In our previous exploration of gene-level splicing analysis, we laid the groundwork for understanding how alternative splicing shapes gene expression. Now, we’re taking a deeper dive into the fascinating world of transcript-level analysis, where we can uncover the intricate details of how genes produce different protein variants through alternative splicing.
//
-

How to Analyze RNAseq Data for Absolute Beginners Part 10: Isoform Analysis
If you’ve followed our previous tutorials on RNA-seq analysis, you’re already familiar with gene-level analysis. But genes are far more complex than simple on/off switches – they can produce multiple versions of RNA transcripts through fascinating processes like alternative splicing. These different versions, called isoforms, allow a single gene to create multiple protein products, dramatically
//
Search
Categories
- bulk RNA-seq (27)
- chromatin accessibility (14)
- Database (4)
- Epigenetics (14)
- Genomics (10)
- HPC (4)
- Metagenomics (1)
- Quick Tips (1)
- RNA-seq (10)
- Scientific Programming (4)
- Single Cell Sequencing (10)
- Transcriptomics (28)
Recent Posts
- How to Analyze Single-Cell RNA-seq Data – Complete Beginner’s Guide Part 7-2: Trajectory Analysis Using Slingshot
- How to Analyze Single-Cell RNA-seq Data from Patient-Derived Xenograft (PDX) Models — Complete Beginner’s Guide Part 8: Processing Human-Mouse Mixed Samples
- How to Analyze Single-Cell RNA-seq Data – Complete Beginner’s Guide Part 7: Trajectory and Pseudotime Analysis Using Monocle 3
- How to Convert BAM Files Back to FASTQ Files: A Practical Guide for NGS Analysis
Tags
Alternative Splicing Analysis ATAC-seq BAM cancer genomics ChIP-seq chromatin accessibility CNV DESeq2 Differential Expression edgeR FASTQ GATK Mutect2 gene expression heatmap HOMER HPC Isoform limma MACS2 MAF miRNA miRNA-seq MSigDB Normalization peak calling RNA-seq somatic mutations Transcript VCF whole genome sequencing



