Category: Transcriptomics
-

How to Analyze RNAseq Data for Absolute Beginners Part 18: Analyzing Viral Gene Expression in Host RNA-seq Data
Understanding viral gene expression patterns during infection is crucial for studying host-pathogen interactions. This comprehensive guide will walk you through the process of accurately quantifying viral transcripts from RNA-seq data of infected host cells, providing you with practical approaches for this challenging analysis. The Challenge of Viral RNA-seq Analysis When we sequence RNA from virus-infected
//
-

How to Analyze RNAseq Data for Absolute Beginners Part 16: A Comprehensive Tutorial on Identifying Fusion Genes
Understanding Fusion Genes: Key Concepts for Cancer Research What Are Fusion Genes and Why Do They Matter? Fusion genes represent a fascinating phenomenon in cancer biology where two previously separate genes join together, often creating proteins with altered or entirely new functions. These genetic mergers typically arise through chromosomal rearrangements like translocations, deletions, or inversions.
//
-

How to Analyze RNAseq Data for Absolute Beginners Part 15-2: Mastering UMI-Based miRNA-Seq Analysis
Understanding UMI-Based miRNA Sequencing MicroRNAs (miRNAs) serve as crucial regulators in gene expression, making their accurate quantification essential for understanding disease mechanisms and biological processes. While traditional miRNA sequencing has proven valuable, the integration of Unique Molecular Identifiers (UMIs) represents a significant advancement in achieving precise miRNA measurements. This tutorial will guide you through the
//
-

How to Analyze RNAseq Data for Absolute Beginners Part 15: A Complete Guide to miRNA-seq Analysis
Understanding the World of microRNAs The fascinating world of microRNAs (miRNAs) represents one of molecular biology’s most elegant regulatory systems. These tiny RNA molecules, spanning just 20-24 nucleotides, function as precise genetic regulators by binding to messenger RNAs (mRNAs) and fine-tuning their expression. Since their serendipitous discovery in the early 1990s, miRNAs have revolutionized our
//
-

How to Analyze RNAseq Data for Absolute Beginners Part 14: Mastering Small RNA-Seq Analysis
The Hidden World of Small RNAs: More Than Just Tiny Molecules Small RNAs are fascinating molecules that challenge the traditional “DNA to RNA to protein” dogma of molecular biology. Despite being just 20-30 nucleotides in length – barely a fraction of a typical messenger RNA – these molecules orchestrate complex biological processes with remarkable precision.
//
-

How to Analyze RNAseq Data for Absolute Beginners Part 13: Circular RNAseq Analysis
Understanding the Biology of Circular RNAs The Nature of Circular RNAs Circular RNAs (circRNAs) represent one of molecular biology’s most fascinating discoveries. Unlike the linear RNA molecules that dominated our understanding of gene expression for decades, circRNAs form continuous loops through a unique process called back-splicing. In this process, a downstream 5′ splice site connects
//
-
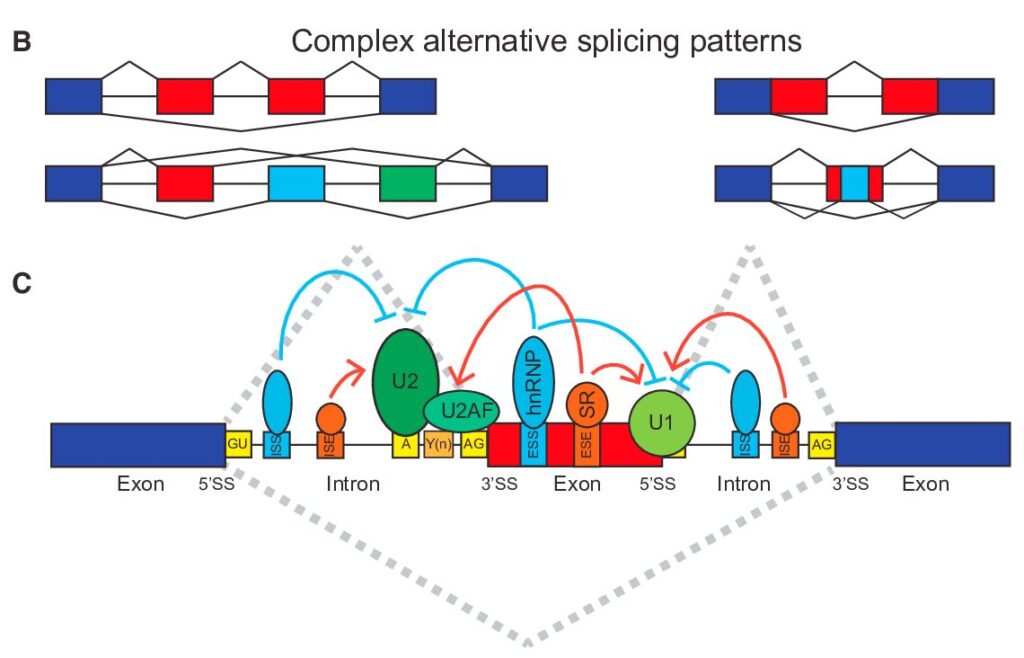
How to Analyze RNAseq Data for Absolute Beginners Part 11: Mastering Transcript-Level Alternative Splicing Analysis
From Gene-Level to Transcript-Level Analysis In our previous exploration of gene-level splicing analysis, we laid the groundwork for understanding how alternative splicing shapes gene expression. Now, we’re taking a deeper dive into the fascinating world of transcript-level analysis, where we can uncover the intricate details of how genes produce different protein variants through alternative splicing.
//
-

How to Analyze RNAseq Data for Absolute Beginners Part 10: Isoform Analysis
If you’ve followed our previous tutorials on RNA-seq analysis, you’re already familiar with gene-level analysis. But genes are far more complex than simple on/off switches – they can produce multiple versions of RNA transcripts through fascinating processes like alternative splicing. These different versions, called isoforms, allow a single gene to create multiple protein products, dramatically
//
-
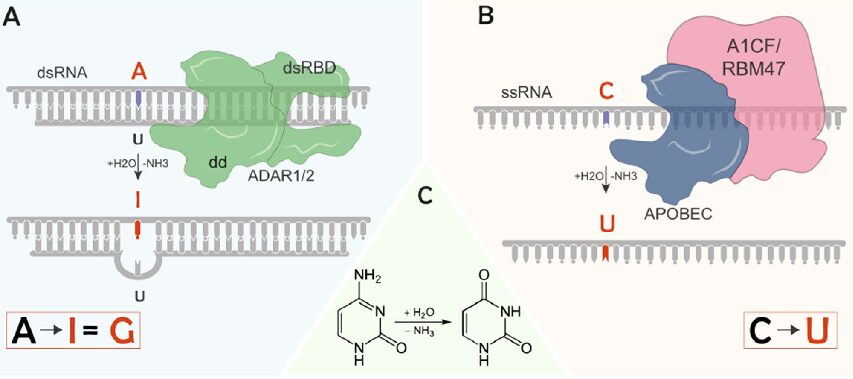
How to Analyze RNAseq Data for Absolute Beginners Part 9: RNA Editing Analysis
RNA editing represents one of the most fascinating mechanisms in molecular biology – a process that introduces targeted changes to RNA sequences after transcription, creating a dynamic transcriptome that diverges from the genome’s blueprint. Through this tutorial, you’ll learn how to analyze RNA editing events from RNA-seq data, uncovering these crucial modifications that fine-tune gene
//
Search
Categories
- bulk RNA-seq (27)
- chromatin accessibility (14)
- Database (4)
- Epigenetics (14)
- Genomics (10)
- HPC (4)
- Metagenomics (1)
- Quick Tips (1)
- RNA-seq (10)
- Scientific Programming (4)
- Single Cell Sequencing (10)
- Transcriptomics (28)
Recent Posts
- How to Analyze Single-Cell RNA-seq Data – Complete Beginner’s Guide Part 7-2: Trajectory Analysis Using Slingshot
- How to Analyze Single-Cell RNA-seq Data from Patient-Derived Xenograft (PDX) Models — Complete Beginner’s Guide Part 8: Processing Human-Mouse Mixed Samples
- How to Analyze Single-Cell RNA-seq Data – Complete Beginner’s Guide Part 7: Trajectory and Pseudotime Analysis Using Monocle 3
- How to Convert BAM Files Back to FASTQ Files: A Practical Guide for NGS Analysis
Tags
Alternative Splicing Analysis ATAC-seq BAM cancer genomics ChIP-seq chromatin accessibility CNV DESeq2 Differential Expression edgeR FASTQ GATK Mutect2 gene expression heatmap HOMER HPC Isoform limma MACS2 MAF miRNA miRNA-seq MSigDB Normalization peak calling RNA-seq somatic mutations Transcript VCF whole genome sequencing




